Measurement, modelling and meta-coordination in cellular communities employing optogenetics tools
Project outline
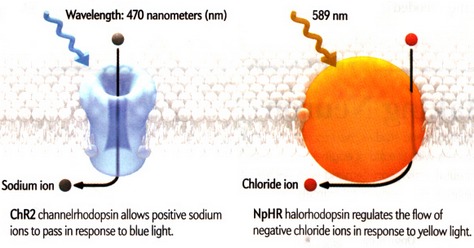
Cells, individually, are complex systems and the behaviour of a cell emerges as a result of molecules interacting in transcriptional, signalling and metabolic networks, as well as interactions with neighbouring cells and the environment. An emerging way to study cellular communities is optogenetics, where targeted illumination is used to control the functions of cells expressing exogenous light-activated proteins. Optogenetics tools allow for precise control of cellular mechanisms and pave the way for a better understanding in biological processes underlying health and disease. The "toolbox" includes opsins, light-sensitive proteins that play photosensory or metabolic roles in species throughout the tree of life; they respond to light of a specific wavelength by transporting or pumping ions into or outside the cells. By genetically engineering different cell lines, the aim of this project is to hijack the cells' own machinery and control ion fluxes through the plasma membrane, monitoring the consequences by the aid of sensors and custom-made optical systems.
Further reading
Optogenetics:Novel Tools for Controlling Mammalian Cell Functions with Light Kushibiki et al. 2014
Optical Developments for Optogenetics Papagiakoumou E. 2013
A Comprehensive Concept of Optogenetics Dugue et al. 2012
Optogenetics-Deisseroth K. Nature Method of the Year 2010